Research
Gene Regulation in Evolution: From Molecular to Extended Phenotypes
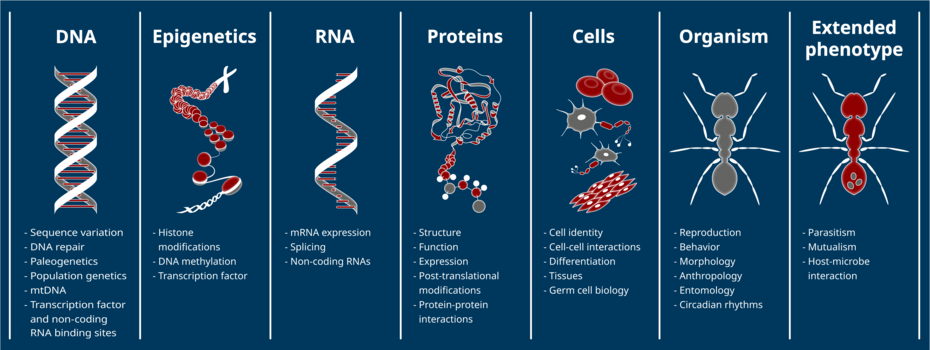
Phenotypic variation between individuals and species is largely driven by differences in gene expression and arises from, among other things, natural selection. Gene expression is regulated via sequence- and non-sequenced-based mechanisms, including cis-regulatory DNA elements, transcription factors, non-coding RNAs, DNA methylation, histone modifications, and other posttranscriptional and posttranslational processes. The relative contribution of these factors in gene regulatory networks as well as the processes underlying their evolution are, so far, poorly understood.
The scientific aim of GenEvo is to gain a better understanding of the evolution of complex and multi-layered gene regulatory systems. We will investigate which regulatory processes are evolutionarily conserved, which are prone to change, and which selective regimes they underlie. By transferring methods developed for model species to other taxa, we will study gene regulatory evolution in a broader intra- and interspecific context. Some projects will even focus on the coevolution of gene regulation, by revealing how interacting species interfere with each other’s gene regulation.
Hint: Projects 1-14 are initial RTG projects. Projects 15-19 have newly emerged during the course of the RTG. Numbers after the decimal point indicate the “generation” of PhD students. Projects marked with "A" belong to associated PhD students. Their research is aligned with the aims of the RTG, but they are funded externally.
Information about individual projects:
Project 1.1 - Mendez DNA methylation inference from archaeological bone: an epigenetic view of human adaptation to selective environments
Project 2.1 - Mulorz The molecular evolution of splicing regulation
Project 3.1 - Poetzsch Gene expression regulation in the adaptive evolution of the hypoxia-tolerant rodent Spalax
Project 4.1 - Elzer Epigenetic information carriers in human sperm cells
Project 5.1 - Govind Comparative analysis of nematode small-RNA pathways using Gametocyte specific factor-1 (GTSF-1)
Project 6.1 - Ceron-Noriega Comparative analysis of evolutionary changes in the transcriptome and proteome in nematodes
Project 7.1 - Scalera Evolution of gene regulation induced by the ultraviolet radiation stress response
Project 8.1 - Conrady Structural Analysis of Monarch butterfly clock proteins
Project 9.1 - Ewerling Evolution of 'cilia' proteins' gene regulatory functions
Project 10.1 - Patterson Epigenetic diversity and adaptation in clonal whiptail lizards
Project 11.1 - Coulm Epigenetic regulation of reproduction in ants
Project 12.1 - Caminer The role of epigenetic mechanisms in the regulation of division of labor in the ant Temnothorax longispinosus
Project 13.1 - Sistermans Parasite interference with the expression of host gene regulation
Project 14.1 - Carvalho Evolution of molecular mechanisms regulating mutualism establishment
Project 1.2 - Tawfik The recent evolution of human life history
Project 2.2 - Hallstein The role of m6A RNA modifications in the evolution of mammalian gene dosage compensation
Project 3.2 - Jelacic Gene expression regulation in the adaptive evolution of the hypoxia-tolerant rodent Spalax
Project 8.2 - Cakilkaya Evolution of BMAL1/CLOCK dependant circadian gene regulation
Project 9.2 - Maißl Co-option of cilia proteins in gene regulatory processes
Project 10.2 - Ku Hybridisation, ploidy, dynamics and reproductive plasticity in whiptail lizards
Project 11.2 - Bolder Role of gene regulation in the social control of queen behavior in ants
Project 12.2 - Knuf Epigenetic regulation of division of labor in the ant Temnothorax longispinosus
Project 13.2 - Blasi Parasite interference in the regulation of host gene expression
Project 15.2 - Carey Investigating adaptation of phase separation to environment in the control of germline specification
Project 16.2 - Amar Emergence of novel regulatory networks from antagonistic interaction between transcription factors and transposable elements
Project 17.2 - Zimmer Evolutionary diversity of sex-chromosome dosage compensation
Project 18.2 - Djolai (née Müller) Investigating the effect of environmental sex determination on gene regulation
Project 19.2 - Wilhelm Evolution of the cellular regulation of a neurotoxin in the Colorado Potato Beetle
Project 1.3 - Mitchell Differential Gene Regulation Across Diverse Human Lifestyles
Project 5.3 - Aaquib Exploring small RNA inheritance as a key mechanism in evolutionary adaptation in C. elegans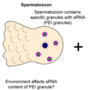
Project 8.3 - Remers Mechanistic insights into the evolution of circadian gene regulationProject 9.3 - Reece Co-option of cilia proteins in gene regulation
Project 10.3 - Priest-Tyson Genetic and Epigenetic Mutation Rates in an Asexually Reproducing Lizard
Project 12.3 - Eberle-Reece The role of epigenetic mechanisms in the regulation of division of labor in the ant Temnothorax longispinosus
Project 13.3 - Chitadze Molecular manipulation of host phenotype via regulatory interference
Project 20.3 - Dominici Sexual dimorphism throughout development in termites
Project 21.3 - Baumgarten Gene regulation and evolution of dormant states in bees
Project 22.3 - Faust The role of small RNas in transgenerational plasticity in a clonal plant
Project 23.3 - Weile Epigenetic adaptation to environmental perturbations
Project 25.3 - Franke Ageing and gene evolution in ants
Project A.01 - Stoldt The transcriptomic basis of adaptive processes in ants of the genus Temnothorax
Project A.02 - Psalti Consistency and molecular mechanisms of ant behaviour
Project A.03 - Ho Allele-specific expression in whiptail lizards
Project A.04 - Collin Genome-wide detection of selection & adaptation in a co-evolving slavemaking ant and its host
Project A.05 - Kanta Epigenetic basis of germline-specific transcription
Project A.06 - Leismann DNA methylation-sensitive transcription factors at the crossroads of transposon and germline genome regulation
Project A.07 - Kalita Evolution of dosage compensation in dipterans
Project A.08 - Bast Mechanisms and evolution of peripheral visual processing strategies
Project A.10 - Lenhart Molecular regulation of division of labor and aging in social insects
Project A.11 - Wagh Cellular and circuit properties that drive differential feature extraction in visual interneurons
Project A.12 - Fraser Sex Determination in Termites
Project A.13 - Michowicz Analysis of Alg13 function during zebrafish germplasm formation
Project A.14 - Agarwal Molecular mechanisms and evolution of neurotoxin tolerance in the Colorado Potato Beetle
Project A.15 - Batista Investigating the molecular mechanisms of transgenerational non-genetic inheritance in Spirodela polyrhiza
Project A.17 - Athreya The co-occurrence of sex and dispersal in Cnidaria and beyond
Project A.18 - Bhat The evolution of ageing as a consequence of limited phenotypic plasticity
Project A.19 - Karimizadeh Consequences of convergent loss of olfactory receptors in slave-making ants
Project A.20 - Hodapp Investigation of the genetic diversity, population health and adaptive potential of solitary bees in Germany
Project A.21 - Srinivasan Theoretical Models of Epigenetic Adaptation Under Geometric Constraints
Project A.22 - Wagner The role of Retinal Pigment Epithelium derived Primary Cilia associated Extracellular Vesicles in Ciliopathy Disease Mechanism
Project A.23 - Mendes The evolution and organization of non-canonical olfactory systems in ants and other insects