Project 22.3
- PhD student: Ida Faust
- Supervisor: Meret Huber
- Co-Supervisors: René Ketting
- Further TAC-members: Arturo Marí-Ordonez
- Research Group
Organisms can alter their phenotype across generations without genetic changes. Yet, the underlying mechanisms remain unclear. It is hypothesized that small RNAs mediate transgenerational gene regulation by priming or retaining stress-induced phenotypes. To investigate the role of small RNAs in transgenerational plasticity, we use the clonal giant duckweed, Spirodela polyrhiza, and its natural herbivore, the water lily aphid, Rhopalosiphum nymphaeae, which appears to manipulate its host plant to its own benefit.
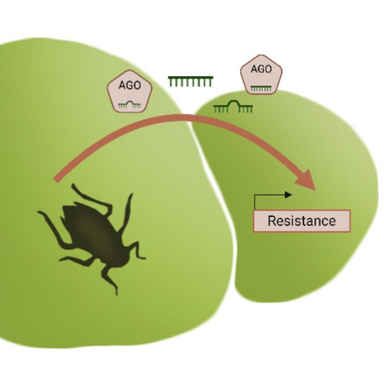
This PhD project investigates the role of small RNAs (sRNAs) as potential molecular carriers of transgenerational information in the aquatic plant Spirodela polyrhiza. S. polyrhiza reproduces clonally, allowing for the precise tracking of stress responses across genetically identical generations. Previous experiments showed that plants were more susceptible to the waterlily aphid when their ancestors, five generations earlier, had been exposed to herbivoresFurthermore, several genes and metabolites were transgenerationally primed. Here, we aim to investigate whether these changes in phenotypes, fitness and gene expression was caused by transgenerationally inherited sRNAs.
sRNAs are known regulators of gene expression and are implicated in transgenerational inheritance in animals. In plants, they are hypothesized to move through the phloem and potentially be passed from mother to offspring. This raises the possibility that sRNAs induced by stress could be inherited across vegetative generations and affect gene expression and plant fitness.
The central aim of this project is to test the hypothesis that herbivory-induced sRNAs are transmitted across clonal generations and shape gene expression and stress responses in S. polyrhiza.
Aphids are sap-feeding insects known to inject saliva into the host-plant while feeding on the phloem. During this close interaction, small molecules like sRNAs could be exchanged and alter gene expression and stress response of the opposite organism. A trans-kingdom, rapid, affordable Purification of RISCs (TraPR) will be used to isolate Argonaut-loaded sRNAs. Mapping these pull-down sRNAs to both the aphid and duckweed genome will help to determine their origin. Further, analysis of DEGs will assess whether sRNas are directly associated with gene expression changes.
The CRISPR/Cas9 system will be applied to generate mutant lines with deficiencies in small RNA biogenesis pathways to investigate whether small RNAs contribute to transgenerational plasticity. Furthermore, candidate sRNAs will be identified and tested for their beneficial effects under stress conditions in a transgenerational stress experiment. Overexpression and knock-out lines will give insights into the role of specific sRNAs.
