Project 8.2
- PhD student: Baris Cakilkaya
- Supervisor: Eva Wolf
- Co-Supervisors: Ann Kathrin Huylmans
- Further TAC-members: Miguel Andrade , Ozkan Yildiz (MPI of Biophysics, Frankfurt)
- Research Group
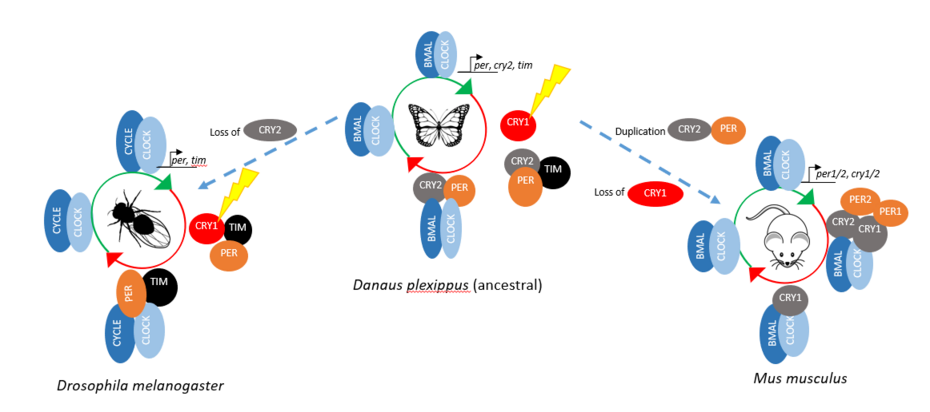
The circadian clock is a gene regulatory mechanism for predicting environmental changes. This mechanism is embedded in each cell, and it is mostly similar in higher organisms with some changes in the negative arm regulations. Danaus plexippus (Monarch butterfly) is a migratory butterfly that contains an ancestral clock machinery. Its machinery has evolved into other insect and mammalian clock architectures. Our aim is to understand this evolution from a structural biologist perspective.
The sun provides energy for life on Earth, and many organisms have adapted to its predictable movement on a daily basis. These organisms have developed an internal mechanism known as the circadian clock, which enables them to adjust their behavior according to the translational oscillation of certain proteins.
The core elements of the circadian clock consist of CLOCK and BMAL transcription factors, which heterodimerize to bind to E-Box elements on the genome, hence they are called activation phase. In activation phase, clock-controlled genes (ccg) are transcribed including repressive proteins of CLOCK:BMAL. Throughout the day, these repressive proteins accumulate and translocate into the nucleus, where they deactivate the CLOCK:BMAL transcription complex, forming the repressive phase. Over the course of night, the repressive protein are degraded, and CLOCK:BMAL initiated transcription resumes, resulting in daily oscillation.
In the repression phase of Danaus plexippus (dp), blue light sensitive CRYPTOCHROME 1 (CRY1) regulates the repression activity by controlling the degradation of TIMELESS (TIM) which is essential for the PERIOD (PER) stability. In the absence of blue light, dpPER and the repressor type dpCRY2 translocate into nucleus to repress dpCLOCK:dpBMAL.
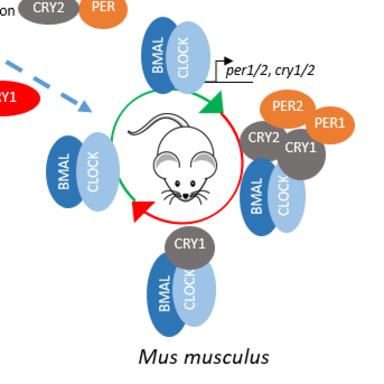
Compared to D. plexippus´s repressor phase, Drosophila melanogaster (Dm) has no repressor type of CRY, and TIM exhibits the repressor activity (Dubowy & Sehgal, 2017). For the mammalian repressor phase, there is no blue light sensitive CRY, however there are two CRYs and PERs, which possess the repression activity. Due to this difference, D. plexippus is considered as the ancient clock which, evolved into D. melanogaster and mammalian.
Interaction of m/dpCRY and BMAL TAD is inhibited by m/dpPER. We hypothesized that this difference is caused by the adjacent region to the CBD of PER. To test the hypothesis, I will be using different dp/mPER constructs to identify the region that competes with BMAL for CRY binding.
The interaction of mCRY1/2 and mCLOCK PAS B is independent from BMAL TAD. mCRY1 shows a stronger binding affinity to CLOCK PAS B than mCRY2, and in the presence of mPER2, mCRY2´s affinity towards mCLOCK PAS B is equalized to mCRY1 (Fribourgh et al., 2020). However, the effect of mPER1 on the interaction of mCRY1/2 and mCLOCK PAS B is unknown. Therefore we aim to investigate the function of mPER1 on mCRY:mCLOCK:mBMAL binding.
Unlike the M. musculus system, our knowledge in the D. plexippus circadian clock machinery is limited. To address this gap in the literature, I will investigate the interaction of dpCRY2 and dpCLOCK PAS B.
References:
Dubowy, C., & Sehgal, A. (2017). Circadian rhythms and sleep in Drosophila melanogaster. Genetics, 205(4). https://doi.org/10.1534/genetics.115.185157
Fribourgh, J. L., Srivastava, A., Sandate, C. R., Michael, A. K., Hsu, P. L., Rakers, C., Nguyen, L. T., Torgrimson, M. R., Parico, G. C. G., Tripathi, S., Zheng, N., Lander, G. C., Hirota, T., Tama, F., & Partch, C. L. (2020). Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. ELife, 9. doi.org/10.7554/eLife.55275