Project A.04
- PhD student: Erwann Collin
- Supervisor: Susanne Foitzik
- Further TAC-members: Joachim Burger, Joseph Colgan , Barbara Feldmeyer (Senckenberg BiK-F)
- Research Group
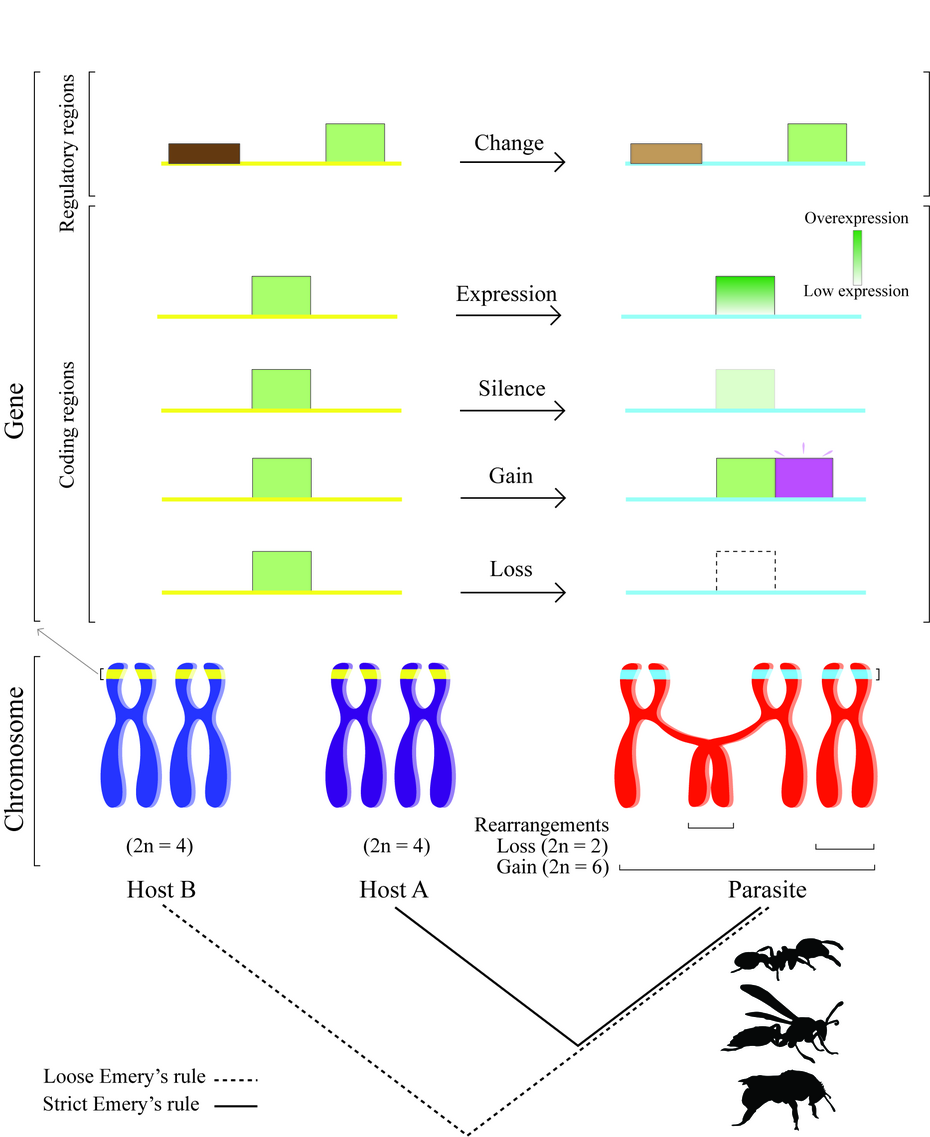
We propose to elucidate the molecular basis of on-going parasite-host coevolution between the slavemaking ant T. americanus and its closely related host species T. longispinosus. We plan to study both sides, the slavemaker and the host, by investigating populations varying in parasite prevalence, and we will place a special focus on genes involved in cuticular hydrocarbon and Dufour gland secretion synthesis and recognition processes.
Coevolution between antagonistic species can drive evolutionary arms races - reciprocal cycles of coadaptation. Recent advances in sequencing technologies allow now to study the molecular basis of evolutionary adaptations on a genomic level in non-model systems. In this project, we focus on a well-studied host-parasite system consisting of the slavemaking ant Temnothorax americanus, an obligate social parasite, and its related host Temnothorax longispinosus, for which ample evidence for coevolution and local adaptation exists. During our 1stfunding phase, we used transcriptomics to investigate, which genes underlie slave raiding and host defense behavior in three slavemaking ants and their three host species of the genus Temnothorax. We detected candidate genes upregulated during slave raids and under selection. First RNAi experiments revealed their potential function in controlling raiding behaviors. We found little overlap between species, both among the differentially expressed genes and those under selection, indicating species-specific evolutionary trajectories. A first population-level project demonstrated that host responses and brain gene expression depends on slavemaker origin, that is parasite prevalence of the slavemaker population. In the 2nd funding phase, we propose to go a step further and investigate the molecular basis of coevolution on a genome-wide level. This is possible as we recently obtained and annotated the genomes of our two main model species. We will use populations of a “natural experiment”, in which host and parasite evolve in sympatry (=coevolve) or allopatry. We propose to study the genetic background of adaptations, the types and strength of selection and whether mainly regulatory regions or protein- coding genes are important for coadaptation. We will use a combination approach of genome-wide Pool-Seq and Resequencing ant genomes from 12 host and 8 parasite populations to determine genes and regions under selection and linkage. We will characterize chemical traits (cuticular hydrocarbons and Dufour gland secretions) and tissue-specific gene expression of all populations to be able conduct a genome-wide association study and link the genomic information to chemical, transcriptomic and behavioral phenotypes. By analyzing multiple populations under similar selective regimes, we can reveal whether coevolution takes different avenues in different locales or whether it occurs in parallel. Once we have identified interesting candidate loci, we will conduct functional RNAi analyses to characterize their effect on the phenotype experimentally. Overall, our study will contribute to a better understanding of genetic mechanisms of adaptation during coevolution.